Researchers Develop Method to Create 3D ‘Map’ of Tissue Structure and Function
Jan 27, 2021 — Atlanta, GA
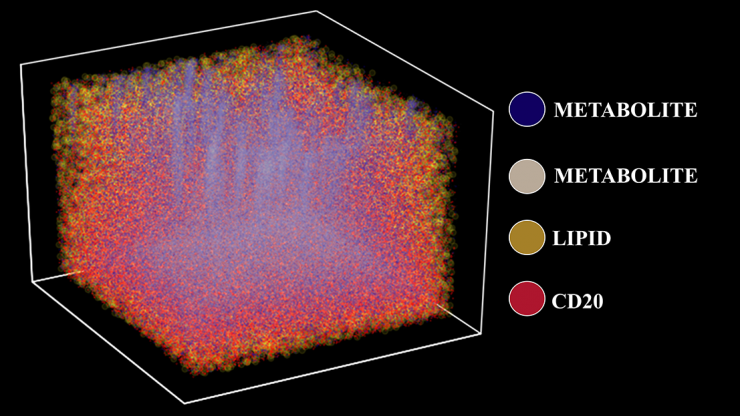
Three-dimensional molecular map of a tonsil using TOF-SIMS, a 3D chemical imaging technique that visualizes metabolic and cellular profiles of more than 190 compounds from a human tissue sample. The plot depicts four distinct chemical and protein signatures — lipids, metabolites, and isotope-labeled cells (CD20). (Image Courtesy: Ahmet Coskun)
A newly published approach to profiling human tissue samples can build a 3D picture of structure and function at the molecular level. The procedure marries techniques from chemistry, biology, and data science and could help doctors design precision therapies in the coming years for patients who aren’t responding to treatment.
In a study of human tonsil tissue, the researchers combined a labeling scheme using isotopes to “tag” specific kinds of cells — in this case, immune cells such as T-cells and B-cells — with imaging mass spectrometry that can identify metabolites, the molecules around those cells that are used for various metabolic functions. And instead of doing this on a single, two-dimensional “slice” of tissue, they used data from about 150 slices to create a 3D map of the tissue.
“An analogy to our system is actually geography: We create the geography of tonsils — where are the valleys, where are the mountains. But when we are doing that, we are looking at more granular features, [including] which molecular distributions are around, and how do they really change within this tonsil tissue,” said Ahmet Coskun, Bernie Marcus Early Career Professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University.
“Typically, the metabolites are measured in one experiment, and these protein-specific labels are measured in another, separate experiment. Bringing the two together in a unique, single measurement is one of the advantages here,” he said.
Coskun’s team used tools from data science to turn all of that data into a 3D map of the tonsil tissue, which Coskun said is more accurate since the tissues are three-dimensional themselves. They described their unique approach — combining two disparate measurements into a single test and processing a huge amount of data to make a 3D map — Jan. 27 in the journal Science Advances. They call the technique a “3D Spatially resolved Metabolomic profiling Framework.”
“You've seen in hospitals how MRIs are done — they can make entire body 3D. That's not done at the microscale, unfortunately,” Coskun said. “When you look at microscale things, they're just two-dimensional slices most of the time.”
Coskun and his team studied B-cells in tonsils, important harbingers of a potential infection. Tonsils are one of the first areas that sense a foreign bacteria or virus, and the immune cells there warn the body to prepare to fight an invader. The team’s spatial map showed the locations of T-cell and B-cell concentrations. It also discovered lower concentrations of specific kinds of fat molecules called lipids that the B-cells use to proliferate and create antibodies.
In experiments looking at nearly 200 different kinds of metabolites and lipids, the researchers uncovered a unique “code” that identified where specific lipids related to different kinds of cell function were depleted.
“That is information you can use to understand how tonsils respond to outside foreign objects that are interfering our bodies,” Coskun said, “and then how those specific anatomical regions in tonsils use their metabolites and lipid content to respond to them.”
Understanding structure and function in conjunction also can pinpoint how cells are using energy, depleting oxygen, or otherwise working in the body, he noted.
“You can use this information to design precision therapies and to find the best drugs for that specific person,” Coskun said. “Drug libraries attack this specific mechanism or that specific mechanism, so by comparing the drug libraries and a specific patient's structure and function profiles, you can actually design personalized drugs for that specific patient.”
That’s still a few years down the road — Coskun said the specialized machines his team used to develop the metabolic profiling are still expensive and mostly housed in research centers.
But: “The biochemical methods that we developed, they're easy; they can be done in any lab,” he said. “Getting the measurements done is the rate-limiting step here.”
Coskun said he could imagine a centralized service where healthcare providers send patient samples for testing — akin to how genetic sequencing is done now. But as technology advances and the machines get cheaper, he said they could end up in more and more hospitals. His team also is developing a cheaper, custom device to overcome the limitations of requiring costly, relatively rare equipment to employ his team’s approach.
“Our goal is discovery. We'd like to use this machine to survey a number of patients who respond to certain drugs and who don't respond to certain drugs,” Coskun said. “We'd like to compare these groups of patients for a personalized therapy approach. We want to make the framework from our end, so that it's ready for clinicians to adapt later.”
Coskun’s team on the study included graduate students Shambavi Ganesh, Thomas Hu, Mayar Allam, and Shuangyi Cai as well as Georgia Tech Institute for Electronics and Nanotechnology researchers Eric Woods and Walter Henderson.
Already, Coskun and his collaborators are exploring what his 3D profiling approach can tell researchers about lung and prostate cancers, visualizing how immunotherapies affect the interplay between the immune system and tumors.
“There are cancer cells and immune cells, and we'd like to understand why and how they're interacting, and then, when they come together, what happens to the structure and function in what we call the tumor microenvironment,” he said.
“If you understand the structure and function of that tumor macroenvironment better, then you can trace back which mechanisms worked or haven't worked.”
This research was supported by the National Institutes of Health K25 Career Development Award K25AI140783, the Burroughs Wellcome Fund, and the Bernie Marcus Early Career Professorship. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of any funding agencies.
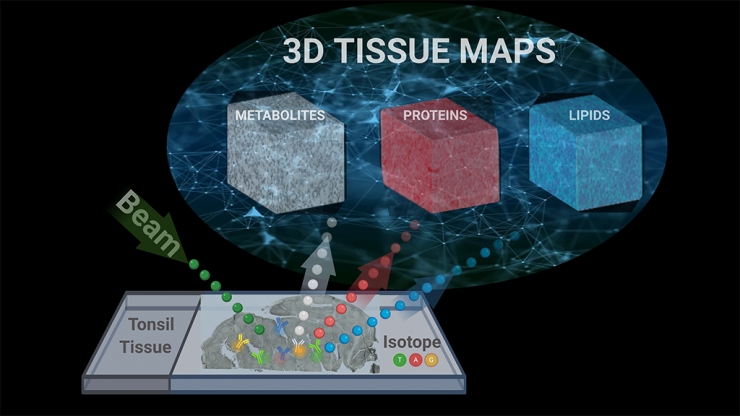
A conceptual diagram illustrates how researchers used a single imagining technique called TOF-SIMS, or time-of-flight secondary ion mass spectrometry, to capture metabolic and cellular profiles of more than 190 compounds in human tissue samples. Ahmet Coskun’s research team used tools from data science to turn all of that data into a 3D map of the tonsil tissue. Their technique is reported in Science Advances. (Image Courtesy: Ahmet Coskun)

Ahmet Coskun, Bernie Marcus Early Career Professor
404.385.2416




